Introduction
A bacterial biofilm is a conglomeration of microorganisms where cells are ensconced within a self-formulated matrix of extracellular polymeric substances, which is primarily composed of polysaccharides, proteins, lipids, and extracellular DNA.1–3 Differing from planktonic bacteria, which can move freely within a bulk solution, cells within a biofilm undergo cell-to-cell interactions. These interactions occur either within the biofilm, with direct contact to the solid surface, or within flocs, where mobile biofilms form without adhering to the surface. The formation of biofilm is an intricate microbial process that encompasses various developmental phases, with some being highly specific to the types of bacteria engaged.4 According to Funari and Shen,3 three basic stages can be distinguished in biofilm formation. In the first stage, adhesion of planktonic bacteria takes place, which is promoted by bacterial appendages such as curli, cilia, and flagella, followed by aggregation and biofilm formation by bacteria. In the second stage, the biofilm grows and matures primarily through the stabilization of the bacteria and interaction with surface which is facilitated by extracellular polymeric substances, enhancing surface adhesion and ensure chemical as well as mechanical protection for the microorganisms. Finally, during the third stage, as the biofilm nears a critical thickness, it initiates the release of bacteria into the surrounding environment to colonize new surfaces. In the course of biofilm formation, microorganisms own the capability to communicate via quorum sensing. This mechanism regulates the metabolic activity of planktonic cells and has the potential to trigger microbial biofilm formation along with heightened virulence.5 In contrast to free-living bacterial cells, extracellular polymeric substances can establish a distinctive local microenvironment that serves various functions, including enhanced resource uptake and surface adhesion, intercellular interactions, and horizontal gene transfer. The matrix offers digestion capacity; protection from external factors such as antibiotics,6 disinfectants,7 and dynamic environments;8 and inhibition of bacterial dehydration.9 Furthermore, the gradient of nutrients and bacterial metabolites in the biofilm results in areas where cells are in a “dormant” state, which means that the cells have extremely reduced metabolic activity.10 These dormant cells are highly resistant to traditional antibiotics, that typically focus on growing and metabolically active. Consequently, addressing bacterial biofilms demands antibiotic doses that are hundreds or even thousands of times higher than those needed to eradicate planktonic forms.3,11 The situation is all the more serious and complicated because, according to Dalton and March,12 approximately 99% of the global bacterial population exists in the form of biofilms, manifesting at different stages of growth. Recent strategies for controlling biofilms, implemented in both hospitals and the food industry (such as cleaning, disinfection, and surface preconditioning), demonstrate some degree of efficacy.13 Nevertheless, their effectiveness falls short of the desired outcomes, and biofilm-induced infections frequently experience recurrence. Hence, there is a need for novel strategies to target bacterial biofilms. One potential option is the use of natural agents, such as farnesol, against bacterial biofilm formation.
Farnesol, a sesquiterpene alcohol, is present in essential oils and is a hydrophobic compound extracted from various sources such as lemongrass, citronella, cyclamen, rose, balsam, neroli, and musk.14–16 It can be endogenously synthesized by dephosphorylating farnesyl pyrophosphate as part of the cholesterol biosynthesis pathway.17,18 Farnesol possesses the capacity to impede the growth of medically significant pathogens,19 as acknowledged by its safety designation from the Food and Drug Administration (USA) as well as by the European Chemicals Agency. This compound impacts the cell membranes of various bacterial species, leading to ion imbalance, ion leakage, and ultimately, cellular death. Farnesol can also interfere with the rate of glycan synthesis in the biofilm matrix, impairing biofilm establishment.20,21 For example, in Streptococcus mutans, momentary exposure to farnesol influenced the growth and metabolism of bacteria by disturbing the bacterial membrane and elevating the polysaccharide content within streptococcal biofilms.20 Studies focused on the mode of action of terpenic alcohols (including farnesol) on Staphylococcus aureus suggested that cell membrane damage may be one of the main antibacterial mechanisms. The leakage rate of K+ ions in bacterial suspensions increased in the presence of farnesol, suggesting that it has antibacterial activity.22 Moreover, studies demonstrate an antifungal effect of this compound by preventing biofilm formation.23 When applied externally, farnesol—tested on a Candida albicans strain lacking endogenous farnesol production—undermines hyphal formation by causing morphological alterations in the yeast cell wall and suppressing the expression of aspartyl proteinases.24 In spite of the encouraging therapeutic effectiveness demonstrated by farnesol against bacterial cells and their biofilms, its poor retention and hydrophobicity limit its use in practice.25
Nanoparticles are objects that in all three dimensions range from 1 to 100 nm. Because of their small size, nanoparticles have different magnetic, optical, and electronic properties (compared with macroscopic materials such as dust, aggregates, and particles) and thus possess different physical and chemical properties. The properties of nanoparticles are related to their size and shape. Nanoparticles have a higher surface-to-volume ratio, and they can interact with bacteria at many levels. Among other things, they act at the membrane level by destabilizing and damaging coatings. The result is increased membrane permeability, leakage of cytoplasm, and subsequent cell death. It has also been shown that nanoparticles can interact with cell wall proteins that include sulfur. By combining with sulfur, nanoparticles cause structural damage, leading to the disruption of the cell wall.26,27 Nanoparticles can penetrate the interior of a bacterial cell and then combine with proteins and nucleic acids. Once combined, the structure of the molecules, and thus their function, is altered. In the same way, nanoparticles can damage genetic material and elements of the respiratory chain, eg, by binding to ATPase and impairing its function. The accumulation of reactive oxygen species through the interaction of nanoparticles with cellular enzymes can lead to further damage inside the cell.28,29 Some nanoparticles (eg, silver, copper) have the ability to inhibit the formation of biofilms by bacteria, thereby increasing the exposure of bacteria to antibacterial agents.30 Efflux pumps also contribute significantly to excluding or including quorum-sensing biomolecules responsible for biofilm formation by bacterial cells. The transit of quorum-sensing biomolecules into or out of bacterial cells can be disrupted by impeding the functionality of efflux pumps. Metallic nanoparticles might block efflux pumps in bacterial cells, thus preventing active pumping of nanoparticles out of the cytoplasm.31 Generally, metal-based nanoparticles, such as silver nanoparticles, copper nanoparticles, zinc oxide nanoparticles, or gold nanoparticles, are widely used in the biomedical sciences. Due to their well-described antimicrobial activity against Gram-positive and Gram-negative bacteria and their antifungal features, these particles offer an alternative to traditionally used antibiotics.32 Considering the antimicrobial properties of metal nanoparticles and farnesol, we hypothesized that biocomponents of metal nanoparticles and farnesol may have the potential to be used as coadjuvants in therapies against bacterial or fungal infections. Thus, the aim of the study was to evaluate the effect of nanocomposites consisting of metal/metal oxide nanoparticles with farnesol on both bacterial and yeast species. We hypothesized that metal/metal oxide nanoparticles damage individual cells, whereas farnesol negatively affects the biofilm’s structure, hindering biofilm formation. Thus, both materials act at two different levels, thereby enhancing each other’s properties.
Materials and Methods
Nanoparticles and Farnesol
Silver (Ag), gold (Au) were obtained from Nano-Koloid (Warsaw, Poland), copper (Cu) from Nano-Tech (Warsaw, Poland) in the form of colloidal water suspensions, and zinc oxide (ZnO) nanopowder was obtained from SkySpring Nanomaterials (Houston, TX, USA). Farnesol (F) (3,7,11-trimethyl-2,6,10-dodecatrien-1-ol, 95%) was purchased from Sigma-Aldrich (Sigma-Aldrich, Hamburg, Germany). Each nanomaterial and farnesol were sonicated at 500 W and 30 kHz for 2 min using a Vibra-Cell™ Ultrasonic Liquid Processor (Sonics & Materials, Newton, CT, USA) before their use in experiments. Nanocomposites were prepared by mixing F with Ag, Au, and Cu, and they were left for 15 min for self-organization.
Microorganisms
Escherichia coli (ATCC 25922), Enterococcus faecalis (ATCC 51299), Staphylococcus aureus (ATCC 25923), Pseudomonas aeruginosa (ATCC 27853), and Candida albicans (ATCC 90028) were obtained from LGC Standards (Teddington, UK) in the form of frozen spore suspensions (in 20% (v/v) glycerol at −20 °C). Before their use in experiments, glycerol was removed by washing with distilled water, and microbial strains were cultured in the following media: tryptic soy agar (TSA) for S. aureus, Luria-Bertani agar (LB) for E. coli, brain heart infusion agar (BHI) for E. faecalis, and yeast nitrogen base (YNB) for C. albicans.
Cell Line
The HFFF2 cell line was obtained from ATCC (American Type Culture Collection, Manassas, VA, USA). The cell line was cultured in DMEM (Dulbecco’s Modified Eagle Medium, Gibco) supplemented with 10% FBS (fetal bovine serum, Thermo Fisher Scientific) and 1% penicillin/streptomycin (Thermo Fisher Scientific) at 37 °C in a humidified atmosphere that contained 5% CO2 in an incubator (NuAire DH AutoFlow CO2 Air-Jacketed Incubator, Plymouth, MA, USA).
Physicochemical Analysis
The zeta potential of the nanomaterials was measured using a Smoluchowski approximation at room temperature (23 °C), followed by a measurement of the hydrodynamic diameter by dynamic light scattering (DLS) with the use of a Zetasizer Nano-ZS ZEN 3600 (Malvern Instruments Ltd., Malvern, UK). The nanoparticles were diluted in ultrapure water to a concentration of 25 µg/mL, and farnesol was diluted to a concentration of 1%. Thereafter, they were sonicated at 500 W and 30 kHz for 2 min using a Vibra-Cell™ Ultrasonic Liquid Processor (Sonics & Materials, Newton, CT, USA).
Moreover, each probe (nanoparticles, farnesol, and complexes) was observed using a transmission electron microscope (TEM, JEM-1220, JEOL, Tokyo, Japan) operated at a voltage of 80 keV. The samples were placed onto TEM grids (3 mm 200 mesh Cu grids, Formvar, Agar Scientific, Stansted, UK), dried, and observed immediately.
Bacteria Viability Assay (XTT)
The viability of the microorganisms after exposure to metal nanoparticles and farnesol (separately and in complexes) was determined by Cell Proliferation Kit II (Cat. No. 11465015001, Merck, Darmstadt, Germany). XTT (sodium 3′-[1-[(phenylamino)-carbony]-3,4-tetrazolium]-bis(4-methoxy-6-nitro) benzene-sulfonic acid hydrate), which is a tetrazolium salt, is reduced to colored formazan by dehydrogenase enzymes in metabolically active cells. Microbial suspensions were prepared in distilled phosphate-buffered saline to 0.5 on the McFarland scale (1.5 × 108 cells/mL) and transferred to a 96-well plate. Then, the nanoparticles were added in the following concentrations: 25, 12.5, 6.25, 3.125, 1.56, and 0.78 µg/mL. Farnesol was added in the following percentages: 5%, 2.5%, 1.25%, 0.625%, 0.3125%, and 0.156%. A combination of the two was also used (nanoparticles 10 µg/mL and farnesol 1%). The prepared plates were incubated for 24 h at 37 °C in a microbiological incubator (NUAire, Plymouth, MN, USA). After incubation, 20 μL of XTT mix was added into each well and incubated for 3 h at 37 °C. Absorbance was measured at 450 nm (with a reference wavelength of 690 nm) using a microplate Elisa reader (Infinite M200, Tecan, Durham, NC, USA). The experiments were repeated at least six times for each group. Cell viability was expressed as a percentage of the control samples, consisting of cells without any substances added.
Analysis of Biofilm Formation
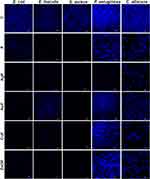
The ability to form a biofilm structure was assessed using DAPI (Sigma-Aldrich, Hamburg, Germany). The microorganisms were placed in an ibiTreat 15 µ-Slide (ibidi GmbH, Gräfelfing, Germany) in 300 µL of growth medium per well containing nanoparticles and farnesol complexes (the final concentrations were 10 µg/mL for nanoparticles and 1% for farnesol). After 48 h, the microorganism cells were washed with PBS three times, and 100 µL of DAPI solution (1 µg/mL) was added for 5 min, protected from light. Thereafter, the solution was removed, and the cells were washed with PBS three times. The formed biofilm was observed with a confocal microscope, FV-1000 (Olympus Corporation, Tokyo, Japan), using a 40× objective at an excitation/emission wavelength of 358/461 nm. Each well was imaged three times, and all images were taken using the same laser parameters.
Analysis of Biofilm Structure
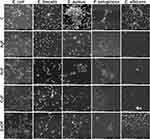
In order to determine changes in biofilm density and changes in microorganism cells, an analysis of structure was performed using scanning electron microscopy (SEM). The microbial cells (3 × 108 cells/mL) were treated with nanoparticles and farnesol complexes (nanoparticles 10 µg/mL with farnesol 1%) and incubated for 48 h at 37 °C in a microbiological incubator (NUAire, Plymouth, MN, USA). Thereafter, the medium with nanoparticles and farnesol was removed, and the cells were washed with PBS three times. The cells were fixed with 2.5% glutaraldehyde (GA) for 30 min at room temperature. Then, the cells were washed with PBS three times and post-fixed with 1% OsO4 in dH2O for 30 min. Subsequently, the cells were washed twice with PBS and dehydrated with a graded ethanol series, as follows: 25% EtOH for 5 min, 50% EtOH for 5 min, 75% EtOH for 5 min, 95% EtOH for 5 min, and 100% anhydrous EtOH for 10 min. The analysis of biofilm structure was performed using SEM (type FEI, Quanta 200, Jeol, Tokyo, Japan) in a low vacuum mode at an accelerating voltage of 20 kV. The pictures of all samples were taken at 10,000× magnification.
Cell Culture Viability
For the evaluation of the viability of the cells exposed to nanoparticles and farnesol, a neutral red (NR, Sigma-Aldrich, Hamburg, Germany) test was performed. HFFF2 cells (1.5 × 105 cells/mL) were seeded on a 96-well plate and incubated for 24 h (37 °C, 5% CO2) so that they could fully attach to the well. Thereafter, nanoparticles and farnesol were added at a concentration of 10 µg/mL and 1%, respectively, for 24 h (37 °C, 5% CO2). Then, 20 µL of NR reagent was added into each well and incubated for 2 h. Absorbance was measured at a wavelength of 570 nm using a microplate Elisa reader (Infinite M200, Tecan, Durham, NC, USA). The experiment was performed at least three times for each group. Viability was expressed as the percentage ratio of the absorbance values of the test sample (cells exposed to nanoparticles and farnesol) to those of the control sample (untreated cells).
Cytokine Array
In order to determine the cytokine expression in HFFF2 cells, a Human Inflammation Antibody Array – Membrane (ab134003, Abcam, Cambridge, UK) for 40 targets was performed. The HFFF2 cells were treated with nanoparticles and farnesol complexes (at concentrations of 10 µg/mL for nanoparticles and 1% for farnesol) and incubated for 24 h. Then, the cells were trypsinized and harvested, and the protein extract was prepared using a TissueLyser LT (Qiagen, Hilden, Germany) at 50 Hz for 10 min with frozen metal balls. After that, the samples were centrifuged at 13,000× g for 10 min at 4 °C, and the pellets were collected. The protein concentration was measured using the BCA Protein Assay (Thermo Scientific) according to the manufacturer’s protocol. The cytokine array was conducted in accordance with the manufacturer’s instructions, and the membranes were imaged using Azure Biosystem C200 (Dublin, CA, USA). The images were then analyzed with ImageJ software (version 1.50e, National Institutes of Health, USA) using the Protein-Array Analyzer plugin.
Statistical Analysis
All the data are presented as mean values ± standard deviation. The data were analyzed with a one-way analysis of variance (ANOVA) with a post hoc Tukey (HSD) test using GraphPad Prism 9 software (version 9.2.0, San Diego, CA, USA). Differences at p ≤ 0.05 were considered statistically significant.
Results
Physicochemical Analysis
Figures 1 and 2 show the values of the average hydrodynamic diameter (in nm) and the zeta potential (in mV) of the materials used in the experiments, respectively. The hydrodynamic diameter of the bare nanoparticles except ZnO (Ag, Au, Cu) was smaller than that in the complexes with farnesol (Table 1). Ag itself had the smallest diameter of all samples. Of all the metal/metal oxide nanoparticles, the ZnO sample had the highest average hydrodynamic diameter (>1 µm), although the size of ZnO indicates the formation of agglomerates, as was also supported by the value of the zeta potential, which was positive (5.0 ± 0.2 mV) and the least different from zero, indicating low colloidal stability. The zeta potential of the other samples was negative. Of the single particles, F had the most negative value (−44.5 ± 0.6 mV), indicating moderate stability of the sample. ZnO, which had a positive zeta potential, showed a highly negative value (−36.1 ± 0.7 mV) after combination with F, indicating a stability similar to that of single F. The zeta potential of Cu, on the other hand, changed from less negative (−7.7 ± 3.3 mV) to more negative (−32.8 ± 3.4 mV) when combined with F. The reduction in stability between Cu and CuF is also evident in Figure 2. The sample with the lowest zeta potential was AgF (−54.1 ± 3.2). The polydispersity index (PdI) fluctuated between 0.3 and 1. AuF and ZnOF had the highest value (1.0), and Ag, Cu, ZnO, and CuF had the lowest value (0.3).
 |
Figure 1 Average hydrodynamic diameter of the materials used in the experiments. (A) Ag; (B) AgF; (C) Au; (D) AuF; (E) Cu; (F) CuF; (G) ZnO; (H) ZnOF; (I) F. |
 |
Figure 2 Zeta potential of the materials used in the experiments. (A) Ag; (B) AgF; (C) Au; (D) AuF; (E) Cu; (F) CuF; (G) ZnO; (H) ZnOF; (I) F. |
The visualization of the materials used is presented in Figure 3. All the metal nanoparticles (Ag, Au, Cu, and ZnO) created agglomerates, with particles clumped together into larger structures. Farnesol, on the other hand, created round droplets with various diameters. The combination of nanoparticles with F resulted in a structure in which the nanoparticles were more separated from each other than without F but in which they formed a denser layer.
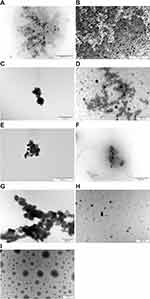 |
Figure 3 TEM visualization of the materials used in the experiments. (A) Ag; (B) AgF; (C) Au; (D) AuF; (E) Cu; (F) CuF; (G) ZnO; (H) ZnOF; (I) F. |
Viability Assay (XTT)
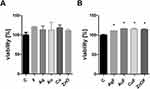
The evaluation of the viability of the microorganisms after F treatment showed decreased viability in most microorganisms except P. aeruginosa (Figure 4). However, F caused the highest percentage of viability reduction in E. faecalis and S. aureus (in both cases, the maximum value of the viability did not exceed 11%). At a concentration of 25 to 6.25 µg/mL, Ag caused statistically significant differences in all microorganism species. Au limited the viability only at concentrations of 25 and 12.5 µg/mL in E. coli and E. faecalis and at the highest concentration (25 µg/mL) in P. aeruginosa and S. aureus and C. albicans. Cu significantly limited the viability at all concentrations in all microorganism strains. ZnO caused a decrease in cell viability of less than 50% in E. coli, E. faecalis, and S. aureus. The complexes AgF, CuF, and ZnOF resulted in a significant decrease in viability in all microorganism strains.
Analysis of Biofilm Formation
Biofilm formation of all microbial species is presented in Figure 5. Changes in biofilm formation after F treatment were especially visible in E. faecalis and S. aureus. In the three other microbial species (E. coli, P. aeruginosa, and C. albicans), biofilm formation was also disturbed, but to a lower extent than in the abovementioned species. For E. coli, AgF and CuF were the factors that most hindered the creation of a biofilm structure. ZnOF contributed to the thinning of the structure, although clustered cells were present. For E. faecalis, most nanocomposites significantly reduced the ability to create a biofilm structure; AuF was the only one that enabled biofilm formation.
For C. albicans and S. aureus, changes in biofilm formation were visible in all nanocomposite samples with F. However, the cells were arranged in clusters, without being divided into individual cells, despite the limited quantitative structure they exhibited in the aforementioned cases. AuF was the least restrictive factor, and therefore the most developed structure was created in its presence. Reduced biofilm formation by S. aureus was observed after exposure to all substances. Only single cells were visible, which did not form a spatial structure. Limited formation of a biofilm, in which only a few cells were present, was noticed for F, AgF, and CuF, while for AuF and ZnOF, the number of cells was slightly higher.
Interestingly, reduced biofilm formation by P. aeruginosa was observed after AgF exposure and to a lesser extent after F treatment. AuF contributed to a biofilm with a flatter shape. CuF and ZnOF treatment resulted in a slightly thinned structure, albeit to a small degree, compared with the control sample.
Analysis of Biofilm Structure
Figure 6 shows representative SEM images for all tested species after exposure to nanocomposites (AgF, AuF, CuF, ZnOF) in the presence of the control group (untreated cells).
For E. coli, the number of bacteria treated with nanocomposites was comparable to that in the control, but their appearance was different in the other samples. Cells exposed to nanocomposites were flattish, with visible cell wall defects. Even though partially degraded, the cells were still arranged in groups.
E. faecalis cells treated with nanocomposites were more separated without a spatial structure (except CuF). Both AuF and AgF caused the cells to be much more flattened, making them appear more elongated than in the control, in which they were arranged in characteristic short chains. In the case of CuF and ZnOF, E. faecalis cells were combined in pairs or small aggregates, although after exposure to CuF, their cell walls were not as smooth and intact as in the ZnOF sample.
For S. aureus, the addition of each nanocomposite reduced the biofilm structure, dividing it into single or slightly merged cells. Only after AuF exposure were the cells still organized in clusters; however, there were fewer clusters than in the control sample, and the cells were more flattened. In all other cases, the cells were partially damaged, with evidence of cell wall rupture and collapse as well as leaking out of intracellular components. After exposure to the nanocomposites, the destroyed structure was less three-dimensional, which, in contrast, was a noticeable feature in the control.
Like E. coli cells, P.aeruginosa cells treated with nanocomposites were more flattened. In the control sample, the cells were surrounded by a layer of extracellular matrix, which was not observed in the samples. The addition of AgF led to cell degradation, even though the cells formed clusters. More biofilm fragments were observed in the AuF sample, although there were degraded cells in the spatial forms as well. Cells exposed to CuF were shrunken, with a collapsed cell wall, mostly not resembling the shape of P. aeruginosa. Similarly shrunken and damaged cells were visible after ZnOF treatment, except that they were still arranged in multidimensional structures.
C. albicans treated with nanocomposites showed a partially reduced structure compared with the control, but the cells were not degraded in all samples. AgF contributed to a significant reduction in the number and morphology of the remaining individual cells that were destroyed. In contrast, AuF did not result in significant cell defects, despite the reduced number of cells present. A reduced number of cells was observed after CuF treatment, and the cells were stacked individually, albeit numerous cells were combined into clusters after ZnOF treatment; however, some cells were destroyed.
Cell Culture Viability
A decrease in cell viability was not observed after exposure to any type of nanocomplex (Figure 7). The HFFF2 viability increased statistically significantly when the cells were treated with AgF, AuF, CuF, and ZnOF. Interestingly, the metabolic activity of the cells did not increase during the treatment with the individual components of the complexes (F, Ag, Au, Cu, ZnO) (Figure 7A).
Cytokine Membrane Assay
Protein expression was analyzed using 40 targets: eotaxin, eotaxin-2, GCSF, GM-CSF, ICAM-1, IFN-γ, I-309, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-6, IL-6sR, IL-7, IL-8, IL-10, IL-11, IL-12p40, IL-12p70, IL-13, IL-15, IL-16, IL-17, IP-10, MCP-1, MCP-2, M-CSF, MIG, MIP-1α, MIP-1β, MIP-1δ, RANTES, TGF-β1, TNF-α, TNF-β, sTNF RI, sTNF-RII, PDGF-BB, and TIMP-2. The results are shown in Figure 8. In the HFFF2 cell line, an increase in expression of the following proteins was noticed: IL-8, MIP-1β, RANTES, and TIMP-2. The expression of the aforementioned proteins increased the most for the groups treated with AgF and ZnOF.
Discussion
Antibiotics were once the most effective agents for combatting potentially lethal diseases. However, such efficient factors are associated with certain side effects that significantly affect their original effectiveness. Unfortunately, the incorrect use of important remedies leads to changes in the functioning of bacteria, which over time adapt to external agents. For this reason, the main problem in antibiotic therapy is antibacterial resistance. The causes of antibiotic resistance vary and include the overuse of antibiotics in agriculture and animal husbandry and the inappropriate prescription and use of antibiotics in medicine. This necessitates the introduction of new or additional agents with antimicrobial activity. Nanoparticles seem to be good candidates for antibacterial agents. Their small size facilitates penetration through the cell wall and membrane of microorganisms, and the large active surface enables the addition of active substances. In this work, we have investigated the influence of complexes of Ag, Au, Cu, and ZnO nanoparticles with farnesol on the viability of some microorganisms (E. coli, E. faecalis, S. aureus, P. aeruginosa, and C. albicans) and biofilm formation. Additionally, the toxicity of the complexes for HFFF2 line fibroblasts was tested.
Analysis of the nanomaterials used is required when performing toxicological studies, as the properties that nanoparticles possess affect their biological effect. Metal nanoparticles tend to agglomerate once they are placed in solution, and this in turn impacts their properties.33 In our study, the hydrodynamic diameter of each type of nanoparticle used (Ag, Au, Cu, ZnO) exceeded 100 nm (Table 1). The formed agglomerates were visible by transmission electron microscopy (Figure 3), although individual nanoparticles had much smaller sizes than the analysis of the average hydrodynamic diameter indicated. However, after combining the nanoparticles with farnesol, an increase in the hydrodynamic diameter was observed for most nanoparticles (Ag, Au, and Cu). Farnesol itself formed particles of various sizes, which were visible during the TEM analysis (Figure 3). Regarding the polydispersity index (PdI), which describes the inhomogeneity of the distribution of particle sizes, the values of the samples ranged from 0.3 to 1.0. A value of PdI exceeding 0.7 means that the sample should not be analyzed by dynamic light scattering because of the broad size distribution of the particles.34 All metal nanoparticles showed a PdI in the range of 0.1 to 0.4, which means that the samples were moderately polydisperse.35 Metal nanoparticles with farnesol had a higher PdI than bare nanoparticles, which may be attributed to the large variety of particles that F exhibited itself. Despite the polydispersity of the complexes with farnesol, the value of the zeta potential increased significantly, surpassing the limit value of ± 30 mV, which is recognized as indicating good colloidal stability of nanoparticles in suspension. This is particularly important in biomedical applications, as unpredictable processes that pose a risk are often the result of poor stability.36 In comparison, none of the metal nanoparticles used alone showed a zeta potential exceeding ± 30 mV (Table 1), although F was a stable factor despite its diverse particle sizes (zeta potential = 44.5 ± 0.6 mV). It can therefore be claimed that the two components (farnesol and metal/metal oxide nanoparticles) are mutually stable, which is particularly important because nanoparticles combine easily with other components owing to their high reactivity surface.36
Metal nanoparticles caused the largest drop in bacterial viability, especially in E. faecalis and S. aureus, which were the two bacterial species that were the most susceptible when treated with Ag, ZnO, and Cu (along with E. coli in this case) (Figure 4). In general, it is believed that Gram-positive bacteria are more resistant to metal nanoparticles than Gram-negative bacteria due to the structure of their cell wall. The wall of Gram-positive bacteria is several times thicker than that of Gram-negative bacteria and therefore may constitute an additional protective barrier.37 However, according to the priority list of pathogens of the World Health Organization (WHO), Gram-negative bacteria are more resistant to antibiotics than Gram-positive bacteria. This is caused by the outer membrane of Gram-negative bacteria, which possess some properties, such as hydrophobicity, that contribute to reduced sensitivity and, consequently, better bacterial resistance.38 In our study, the reaction of Gram-negative bacteria to the exposure to nanoparticles may be caused by the presence of an outer layer with LPS molecules, which increase the negative charge of the whole cell membrane and repel nanoparticles due to their mutual interaction, especially since some regions in the mosaic structure of the LPS molecules in the outer membrane may be more negatively charged.39 Interestingly, C. albicans demonstrated higher sensitivity to metal nanoparticles than Gram-negative bacteria species, but its viability was significantly decreased only in the highest concentrations of nanoparticles tested. The fungal cell wall is composed of substances different from those found in bacteria, eg, glucan, chitin, chitosan, and glycoproteins, which are responsible for its rigidity.40 The effectiveness of metal nanoparticles against C. albicans has nevertheless been proven by showing, among other things, that they attach to the cell wall and membrane and accumulate inside both structures, causing ion release or dysfunction of enzymes and DNA.41 A similar action is observed when nanoparticles interact with bacteria. Exposure of nanoparticles to bacterial cells results in the adhesion of metal nanoparticles to the cell surface, which leads to damage to the cell membrane and penetration of the nanoparticles into the cell. It has already been demonstrated that metal nanomaterials can interact physically with the cell membrane/wall or with other intracellular components.42 Adsorption of nanoparticles on the cell membrane disrupts the transfer of electrons and increases the membrane’s permeability. Nanoparticles are constantly undergoing dissolution in solutions, and the released ions can affect cellular structures. Nanoparticles that interact with the bacterial surface produce a concentrated source of ions, continuously releasing ions and causing more toxicity to the cells. The mechanism of the antimicrobial action of metal ions is closely related to their interaction with sulfhydryl groups in enzymes and proteins, resulting in cell dysfunction.37,43 Metal nanoparticles are well known for their high capacity to produce reactive oxygen species (ROS) and free radicals, the excessive production of which is responsible for lipid oxidation and protein and DNA damage.44 Such a multifaceted interaction of metal nanoparticles with bacterial cells provides many opportunities for their use as antimicrobial agents.
F was a factor that significantly reduced microorganism viability (up to a maximum value of 11%) in two species, namely E. faecalis and S. aureus, in all concentrations used (Figure 4). It also reduced the viability of the yeast C. albicans, but to a smaller extent. As was mentioned in the Introduction, farnesol is recognized as an agent able to inhibit a broad spectrum of relevant pathogens, and its antibacterial properties have already been proven against both E. faecalis and S. aureus.45–47 A different situation is observed in the case of C. albicans, since farnesol is a fungal quorum-sensing molecule; however, when added in the form of an exogenous agent, it contributes to cell wall changes, meaning that it may also be useful as an antifungal compound.24 Quorum sensing is closely related to the formation of a biofilm, which is the spatial three-dimensional structure that most microorganisms form in the environment. Bacteria embedded in a biofilm are able to tolerate 10–1000 times higher concentrations of bactericides and fungicides than planktonic forms of cells.48 Therefore, biofilm formation was limited for C. albicans, but also for the bacterial species used, both during exposure to F alone and during exposure to nanocomposites, especially AgF, with additional toxicity exhibited by Ag, which is known for its antibacterial properties (Figure 5). As mentioned before, metal nanoparticles may be broad-spectrum antibacterial and antifungal agents, destroying both bacterial and yeast cells, meaning that biofilm formation during exposure to nanocomposites may have caused damage within individual cells. In general, most bacteria species produce a negatively charged matrix, so positively charged nanoparticles can easily penetrate it.49 In our study, most nanoparticles, as well as all nanocomposites with F, had a negative charge (except ZnO), suggesting that nanocomposites can better penetrate the biofilm structure, especially because hydrophobic particles (such as F) exhibit a better distribution within biofilm structures than hydrophilic particles.50 Such a distribution of nanocomposites may explain the reduction in biofilm structure, which was observed in the SEM analysis (Figure 6). Moreover, the SEM analysis showed that individual cells, in spite of their limited number, were damaged and had a shape that differed from that of the control samples. In all microorganism samples exposed to AgF, there were fewer cells (or they were separated, as in C. albicans), and they seemed to be more flattened. In this case, the cells did not form clusters, but they existed as individual cells that were not merged together, as opposed to the samples after AuF exposure, in which there were also fewer cells, but they were clumped. Biofilm damage can be caused by the action of F; however, the molecular mechanism of farnesol is not completely clear.51 Interestingly, farnesol may inhibit quorum-sensing molecules such as 2-heptyl-3-hydroxy-4-quinolone in P. aeruginosa, known as the Pseudomonas quinolone signal (PQS). As PQS regulates the synthesis of eDNA (extracellular DNA), which in turn is responsible for cell-to-cell communication and maintenance of the biofilm structure, inhibition of PQS production results in the formation of weak and thin biofilms.52 Such a phenomenon may have occurred in studies that found an arrangement of individual rather than clustered cells. Other research in which the biofilm structure was treated with farnesol and antibiotics, eg, β-lactams, suggested that farnesol interferes with peptidoglycan biosynthesis and, as a consequence, cell wall formation, enhancing the effect of β-lactams.53 Such an effect seemed to be possible in our study, especially as metal nanoparticles may contribute to cell wall damage.
When testing substances with general applicability, it is extremely important to examine the toxicity that the new substance may cause to cells with proper morphology. Farnesol is considered safe by the Food and Drug Administration (USA) as well as by the European Chemicals Agency; however, its effect depends on the concentration: the higher the concentration of farnesol, the greater the damage to cells. There are multiple studies in which farnesol affects cancer cells but, at the same time, does not exhibit toxicity toward healthy cell lines.54 In our research, farnesol with nanoparticles did not show toxicity toward the HFFF2 cell line (Figure 7); however, there was increased expression of cytokines, such as IL-8, MIP-1β, RANTES, and TIMP-2 (Figure 8), especially after treatment with AgF and ZnOF. The results seem to be probable, as F is considered a contact allergen in humans, as well as a mild irritant for rabbit skin.55 Interestingly, F can be both a pro- and an anti-inflammatory agent. Some reports show that F increased the level of IL-6 in macrophage cells, but it also led to inactivation of the Ras-Raf-ERK1/2 signaling pathway, resulting in suppression of inflammatory gene expression.16 Farnesol may be toxic in the proposed nanocomposites, but metal nanoparticles can also cause undesirable effects in both in vitro and in vivo models. Nevertheless, the effects of nanoparticles are dependent on the various properties that they have, including size, shape, and colloidal stability.56 In our research, the AgF composite had the smallest diameter of all samples, and smaller particles pass more easily through the cell membrane and into the cells.57 A similar situation is observed in ZnOF, which had the second-largest size, although zinc oxide nanoparticles release zinc ions, which alter Zn-dependent intracellular enzymes and transcriptional factors in cells.58 Despite some speculation about the toxicity of the listed substances, each cell line may react differently to each substance. For this reason, it is crucial to conduct research on a larger number of cell lines. An important point, however, is the lack of a reduction in cell viability, which was demonstrated in our study and which gives hope for the potential use of those agents without inducing additional general toxicity.
Conclusion
To summarize, nanocomposites composed of metal/metal oxide nanoparticles (silver, gold, copper, and zinc oxide) and farnesol may constitute an effective and safe antibiofilm agent due to the damaging effect that both compounds have. Firstly, metal/metal oxide nanoparticles may damage microorganism cells by a wide range of effects, including cell wall destruction. Secondly, farnesol may impair quorum sensing, leading to the formation of weaker and thinner biofilms than in favorable conditions. However, because different microorganisms have different reactions, metabolic functions, and cell structures and because each nanomaterial has its characteristic properties, many aspects must be considered when selecting a particular antibiofilm compound.
Abbreviations
Ag, silver nanoparticles; ATCC, American Type Culture Collection; Au, gold nanoparticles; BHI, brain heart infusion agar; Cu, copper nanoparticles; DLS, dynamic light scattering; DMEM, Dulbecco’s Modified Eagle Medium; eDNA, extracellular DNA; F, farnesol; FBS, fetal bovine serum; LB, Luria-Bertani agar; NR, neutral red; PdI, polydispersity index; PQS, Pseudomonas quinolone signal; ROS, reactive oxygen species; SEM, scanning electron microscopy; TEM, transmission electron microscope; TSA, tryptic soy agar; WHO, World Health Organization; XTT, sodium 3′-[1-[(phenylamino)-carbony]-3,4-tetrazolium]-bis(4-methoxy-6-nitro) benzene-sulfonic acid hydrate; YNB, yeast nitrogen base; ZnO, zinc oxide nanoparticles.
Funding
The publication was financed by Science development fund of the Warsaw University of Life Sciences – SGGW.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Maier B. How physical interactions shape bacterial biofilms. Annu Rev Biophys. 2021;50(1):401–417. doi:10.1146/annurev-biophys-062920-063646
2. Wong GCL, Antani JD, Lele PP, et al. Roadmap on emerging concepts in the physical biology of bacterial biofilms: from surface sensing to community formation. Phys Biol. 2021;18(5). doi:10.1088/1478-3975/ABDC0E
3. Funari R, Shen AQ. Detection and characterization of bacterial biofilms and biofilm-based sensors. ACS Sens. 2022;7(2):347–357. doi:10.1021/ACSSENSORS.1C02722/ASSET/IMAGES/LARGE/SE1C02722_0003.JPEG
4. Garrett TR, Bhakoo M, Zhang Z. Bacterial adhesion and biofilms on surfaces. Prog Nat Sci. 2008;18(9):1049–1056. doi:10.1016/j.pnsc.2008.04.001
5. Preda VG, Săndulescu O. Communication is the key: biofilms, quorum sensing, formation and prevention. Discoveries. 2019;7(3):e10. doi:10.15190/d.2019.13
6. Goldberg J. Biofilms and antibiotic resistance: a genetic linkage. Trends Microbiol. 2002;10(6):264. doi:10.1016/S0966-842X(02)02381-8
7. Peng J-S, Tsai W-C, Chou -C-C. Inactivation and removal of Bacillus cereus by sanitizer and detergent. Int J Food Microbiol. 2002;77(1–2):11–18. doi:10.1016/S0168-1605(02)00060-0
8. Chen MJ, Zhang Z, Bott TR. Direct measurement of the adhesive strength of biofilms in pipes by micromanipulation. Biotechnol Tech. 1998;12(12):875–880. doi:10.1023/A:1008805326385/METRICS
9. Flemming H-C, Neu TR, Wozniak DJ. The EPS matrix: the house of biofilm cells. J Bacteriol. 2007;189(22):7945–7947. doi:10.1128/JB.00858-07
10. Kim J, Hahn J-S, Franklin MJ, Stewart PS, Yoon J. Tolerance of dormant and active cells in Pseudomonas aeruginosa PA01 biofilm to antimicrobial agents. J Antimicrob Chemother. 2009;63(1):129–135. doi:10.1093/jac/dkn462
11. Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2(2):114–122. doi:10.1038/nrd1008
12. Dalton HM, March PE. Molecular genetics of bacterial attachment and biofouling. Curr Opin Biotechnol. 1998;9(3):252–255. doi:10.1016/S0958-1669(98)80055-4
13. Carrascosa C, Raheem D, Ramos F, Saraiva A, Raposo A. Microbial biofilms in the food industry—a comprehensive review. Int J Environ Res Public Health. 2021;18(4):2014. doi:10.3390/ijerph18042014
14. Azanchi T, Shafaroodi H, Asgarpanah J. Anticonvulsant activity of Citrus aurantium blossom essential oil (neroli): involvement of the GABAergic system. Nat Prod Commun. 2014;9(11):1615–1618.
15. Krupčík J, Gorovenko R, Špánik I, Sandra P, Armstrong DW. Enantioselective comprehensive two-dimensional gas chromatography. A route to elucidate the authenticity and origin of Rosa damascena Miller essential oils. J Sep Sci. 2015;38(19):3397–3403. doi:10.1002/jssc.201500744
16. Jung Y, Hwang S, Sethi G, Fan L, Arfuso F, Ahn K. Potential anti-inflammatory and anti-cancer properties of farnesol. Molecules. 2018;23(11):2827. doi:10.3390/molecules23112827
17. Joo JH, Jetten AM. Molecular mechanisms involved in farnesol-induced apoptosis. Cancer Lett. 2010;287(2):123–135. doi:10.1016/j.canlet.2009.05.015
18. Sato T, Watanabe T, Mikami T, Matsumoto T. Farnesol, a morphogenetic autoregulatory substance in the dimorphic fungus Candida albicans, inhibits hyphae growth through suppression of a mitogen-activated protein kinase cascade. Biol Pharm Bull. 2004;27(5):751–752. doi:10.1248/bpb.27.751
19. Yar N, Wittman E, Schaut D, Seta F. Effects of farnesol on drug-resistant and non-resistant Candida albicans: implications for cosmetic and pharmaceutical applications. Adv Microbiol. 2020;10(08):383–396. doi:10.4236/aim.2020.108028
20. Koo H. Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol. J Antimicrob Chemother. 2003;52(5):782–789. doi:10.1093/jac/dkg449
21. Ivanova A, Ivanova K, Fiandra L, et al. Antibacterial, antibiofilm, and antiviral farnesol-containing nanoparticles prevent Staphylococcus aureus from drug resistance development. Int J Mol Sci. 2022;23(14):7527. doi:10.3390/ijms23147527
22. Inoue Y, Shiraishi A, Hada T, Hirose K, Hamashima H, Shimada J. The antibacterial effects of terpene alcohols on Staphylococcus aureus and their mode of action. FEMS Microbiol Lett. 2004;237(2):325–331. doi:10.1111/j.1574-6968.2004.tb09714.x
23. Ramage G, Saville SP, Wickes BL, López-Ribot JL. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol. 2002;68(11):5459–5463. doi:10.1128/AEM.68.11.5459-5463.2002/ASSET/962BC315-0B68-447D-98C7-A45C742CEE16/ASSETS/GRAPHIC/AM1120413003.JPEG
24. Decanis N. Farnesol, a fungal quorum-sensing molecule triggers Candida albicans morphological changes by downregulating the expression of different secreted aspartyl proteinase genes. Open Microbiol J. 2011;5(1):119–126. doi:10.2174/1874285801105010119
25. Sims KR, Liu Y, Hwang G, Jung HI, Koo H, Benoit DSW. Enhanced design and formulation of nanoparticles for anti-biofilm drug delivery. Nanoscale. 2019;11(1):219–236. doi:10.1039/C8NR05784B
26. Vimbela G, Ngo S, Fraze C, Yang L, Stout DA. Antibacterial properties and toxicity from metallic nanomaterials [Corrigendum]. Int J Nanomed. 2018;13:6497–6498. doi:10.2147/IJN.S183907
27. Bruna T, Maldonado-Bravo F, Jara P, Caro N. Silver nanoparticles and their antibacterial applications. Int J Mol Sci. 2021;22(13). doi:10.3390/ijms22137202
28. Mammari N, Lamouroux E, Boudier A, Duval RE. Current knowledge on the oxidative-stress-mediated antimicrobial properties of metal-based nanoparticles. Microorg. 2022;10(2):437. doi:10.3390/MICROORGANISMS10020437
29. Ananth A, Dharaneedharan S, Heo MS, Mok YS. Copper oxide nanomaterials: synthesis, characterization and structure-specific antibacterial performance. Chem Eng J. 2015;262:179–188. doi:10.1016/J.CEJ.2014.09.083
30. Lange A, Grzenia A, Wierzbicki M, et al. Silver and copper nanoparticles inhibit biofilm formation by mastitis pathogens. Anim. 2021;11(7):1884. doi:10.3390/ANI11071884
31. Gupta D, Singh A, Khan AU. Nanoparticles as efflux pump and biofilm inhibitor to rejuvenate bactericidal effect of conventional antibiotics. Nanoscale Res Lett. 2017;12. doi:10.1186/S11671-017-2222-6
32. Sánchez-López E, Gomes D, Esteruelas G, et al. Metal-based nanoparticles as antimicrobial agents: an overview. Nanomater. 2020;10(2):292. doi:10.3390/NANO10020292
33. Medici S, Peana M, Pelucelli A, Zoroddu MA. An updated overview on metal nanoparticles toxicity. Semin Cancer Biol. 2021;76:17–26. doi:10.1016/J.SEMCANCER.2021.06.020
34. Danaei M, Dehghankhold M, Ataei S, et al. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharm. 2018;10(2):57. doi:10.3390/PHARMACEUTICS10020057
35. Takechi-Haraya Y, Ohgita T, Demizu Y, Saito H, Izutsu K, Sakai-Kato K. Current status and challenges of analytical methods for evaluation of size and surface modification of nanoparticle-based drug formulations. AAPS Pharm Sci Tech. 2022;23(5):150. doi:10.1208/s12249-022-02303-y
36. Xu L, Liang HW, Yang Y, Yu SH. Stability and reactivity: positive and negative aspects for nanoparticle processing. Chem Rev. 2018;118(7):3209–3250. doi:10.1021/ACS.CHEMREV.7B00208/ASSET/IMAGES/MEDIUM/CR-2017-00208M_0040.GIF
37. Slavin YN, Asnis J, Häfeli UO, Bach H. Metal nanoparticles: understanding the mechanisms behind antibacterial activity. J Nanobiotechnology. 2017;15(1):1–20. doi:10.1186/S12951-017-0308-Z/FIGURES/4
38. Breijyeh Z, Jubeh B, Karaman R. Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Mol. 2020;25(6):1340. doi:10.3390/MOLECULES25061340
39. Franco D, Calabrese G, Guglielmino SPP, Conoci S. Metal-based nanoparticles: antibacterial mechanisms and biomedical application. Microorg. 2022;10(9):1778. doi:10.3390/MICROORGANISMS10091778
40. Garcia-Rubio R, de Oliveira HC, Rivera J, Trevijano-Contador N. The fungal cell wall: candida, Cryptococcus, and Aspergillus species. Front Microbiol. 2020;10:492056. doi:10.3389/FMICB.2019.02993/BIBTEX
41. Carmo PHF, Garcia MT, Figueiredo-Godoi LMA, Lage ACP. Metal nanoparticles to combat Candida albicans infections: an update. Microorg. 2023;11(1):138. doi:10.3390/MICROORGANISMS11010138
42. Dizaj SM, Lotfipour F, Barzegar-Jalali M, Zarrintan MH, Adibkia K. Antimicrobial activity of the metals and metal oxide nanoparticles. Mater Sci Eng C. 2014;44:278–284. doi:10.1016/J.MSEC.2014.08.031
43. Qing Y, Cheng L, Li R, et al. Potential antibacterial mechanism of silver nanoparticles and the optimization of orthopedic implants by advanced modification technologies. Int J Nanomed. 2018;13:3311–3327. doi:10.2147/IJN.S165125
44. Su HL, Chou CC, Hung DJ, et al. The disruption of bacterial membrane integrity through ROS generation induced by nanohybrids of silver and clay. Biomaterials. 2009;30(30):5979–5987. doi:10.1016/J.BIOMATERIALS.2009.07.030
45. Kaneko M, Togashi N, Hamashima H, Hirohara M, Inoue Y. Effect of farnesol on mevalonate pathway of Staphylococcus aureus. J Antibiot. 2011;64(8):547–549. doi:10.1038/ja.2011.49
46. Jabra-Rizk MA, Meiller TF, James CE, Shirtliff ME. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob Agents Chemother. 2006;50(4):1463–1469. doi:10.1128/AAC.50.4.1463-1469.2006
47. De-Carli AD, Sorgatto DL, Paim R, et al. Antimicrobial activity of tt-farnesol associated with an endodontic sealer against Enterococcus faecalis. G Ital Endod. 2021;35(1):10–16. doi:10.32067/GIE.2021.35.01.12
48. Abebe GM. The role of bacterial biofilm in antibiotic resistance and food contamination. Int J Microbiol. 2020;2020:1–10. doi:10.1155/2020/1705814
49. Shkodenko L, Kassirov I, Koshel E. Metal oxide nanoparticles against bacterial biofilms: perspectives and limitations. Microorganisms. 2020;8(10):1545. doi:10.3390/microorganisms8101545
50. Sahli C, Moya SE, Lomas JS, Gravier-Pelletier C, Briandet R, Hémadi M. Recent advances in nanotechnology for eradicating bacterial biofilm. Theranostics. 2022;12(5):2383–2405. doi:10.7150/thno.67296
51. Chen S, Xia J, Li C, Zuo L, Wei X. The possible molecular mechanisms of farnesol on the antifungal resistance of C. albicans biofilms: the regulation of CYR1 and PDE2. BMC Microbiol. 2018;18(1):1–14. doi:10.1186/S12866-018-1344-Z/TABLES/5
52. Bandara HMHN, Herpin MJ, Kolacny D, Harb A, Romanovicz D, Smyth HDC. Incorporation of farnesol significantly increases the efficacy of liposomal ciprofloxacin against Pseudomonas aeruginosa biofilms in vitro. Mol Pharm. 2016;13(8):2760–2770. doi:10.1021/ACS.MOLPHARMACEUT.6B00360/ASSET/IMAGES/LARGE/MP-2016-00360P_0008.JPEG
53. Castelo-Branco DSCM, Riello GB, Vasconcelos DC, et al. Farnesol increases the susceptibility of Burkholderia pseudomallei biofilm to antimicrobials used to treat melioidosis. J Appl Microbiol. 2016;120(3):600–606. doi:10.1111/jam.13027
54. Öztürk B Y, Feyzullazade N, İ D, Şengel T. The investigation of in vitro effects of farnesol at different cancer cell lines. Microsc Res Tech. 2022;85(8):2760–2775. doi:10.1002/jemt.24125
55. Farnesol | C15H26O | CID 3327 – PubChem. Available from: https://pubchem.ncbi.nlm.nih.gov/compound/Farnesol.
56. Zhang N, Xiong G, Liu Z. Toxicity of metal-based nanoparticles: challenges in the nano era. Front Bioeng Biotechnol. 2022;10:1001572. doi:10.3389/FBIOE.2022.1001572/BIBTEX
57. Sukhanova A, Bozrova S, Sokolov P, Berestovoy M, Karaulov A, Nabiev I. Dependence of nanoparticle toxicity on their physical and chemical properties. Nanoscale Res Lett. 2018;13(1):44. doi:10.1186/s11671-018-2457-x
58. R VG, M PV. Cellular interactions of zinc oxide nanoparticles with human embryonic kidney (HEK 293) cells. Colloids Surf B Biointerfaces. 2017;157:182–190. doi:10.1016/j.colsurfb.2017.05.069